Bespoke Software for Omics
We build modern scientific software — from cloud pipelines to custom dashboards — that turns your raw proteomics data into results.

🗄️ Scientific Databases
- We design Django + PostgreSQL schemas for peptides, proteins & metadata.
- Our solutions include fast search and secure REST / GraphQL APIs.
- We deliver built-in charts and export formats (Skyline, CSV, JSON).
Live demo: sysquan.com
Let’s design your database →🛠️ Cloud Pipelines & Servers
- We deploy containerised workflows on AWS, GCP, Hetzner or on-prem.
- Setups include automation via Snakemake, Nextflow or Bash.
- We also offer 24/7 monitoring, backups and secure hand-off.

📈 ML & QC Models
- We offer peptide rescoring, contaminant detection, and RT prediction.
- Models are built using SVMs, boosting trees or deep learning.
- You receive a Python package or deployable REST microservice.
Latest work: Nature Comms – MHCvalidator
Scope an ML project →🌐 Interactive Dashboards
- We build dashboards with Streamlit, React or Django templates.
- Applications include QC charts, SOP navigation and filtering tools.
- All apps support sign-in via Google or Azure with full audit tracking.
Example: MhcVizPipe QC tool
Preview UI ideas →
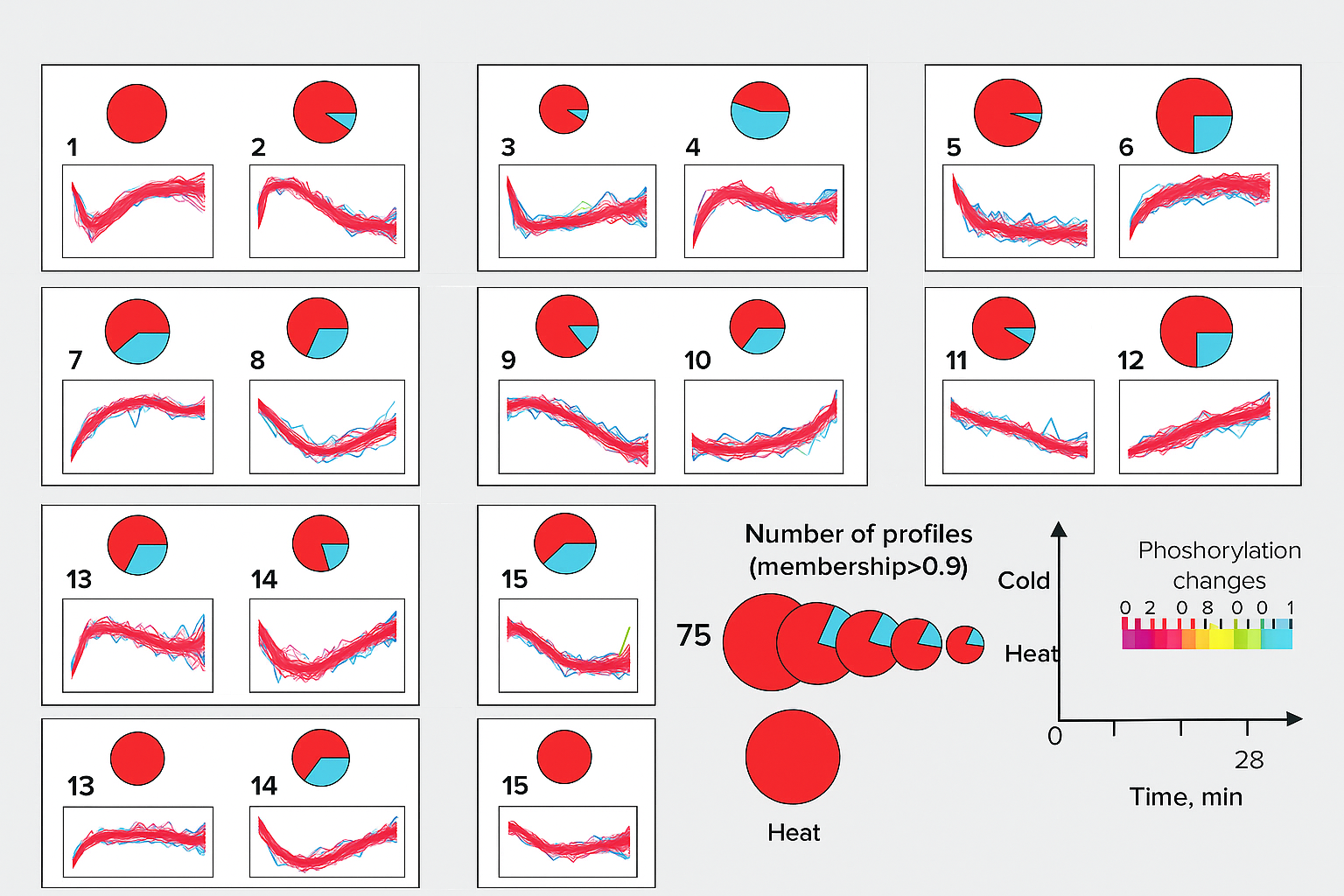
🧬 Bioinformatics Pipelines
- We help you go from raw MS files → IDs → quant → stats → plots.
- Support for PTM analysis, time-course clustering and enrichment.
- Each pipeline is packaged in Conda or Docker for long-term use.
Method used in our yeast stress phosphoproteome study.
Get a pipeline quote →